Single Cell Sequencing using DropSeq
Both by request of Dr. Leiboff, and inspired by the elegant work of Shalan et al. 2020 (https://www.biorxiv.org/content/10.1101/2020.06.29.178863v1), I animated this series to illustrate the basic principles of single cell sequencing using microfluidic droplets.
These animations were used in a Intro Genetics class, and I will continue to improve upon them for the community to use for educational purposes.

The root apical meristem is highly structured with specific cell identities
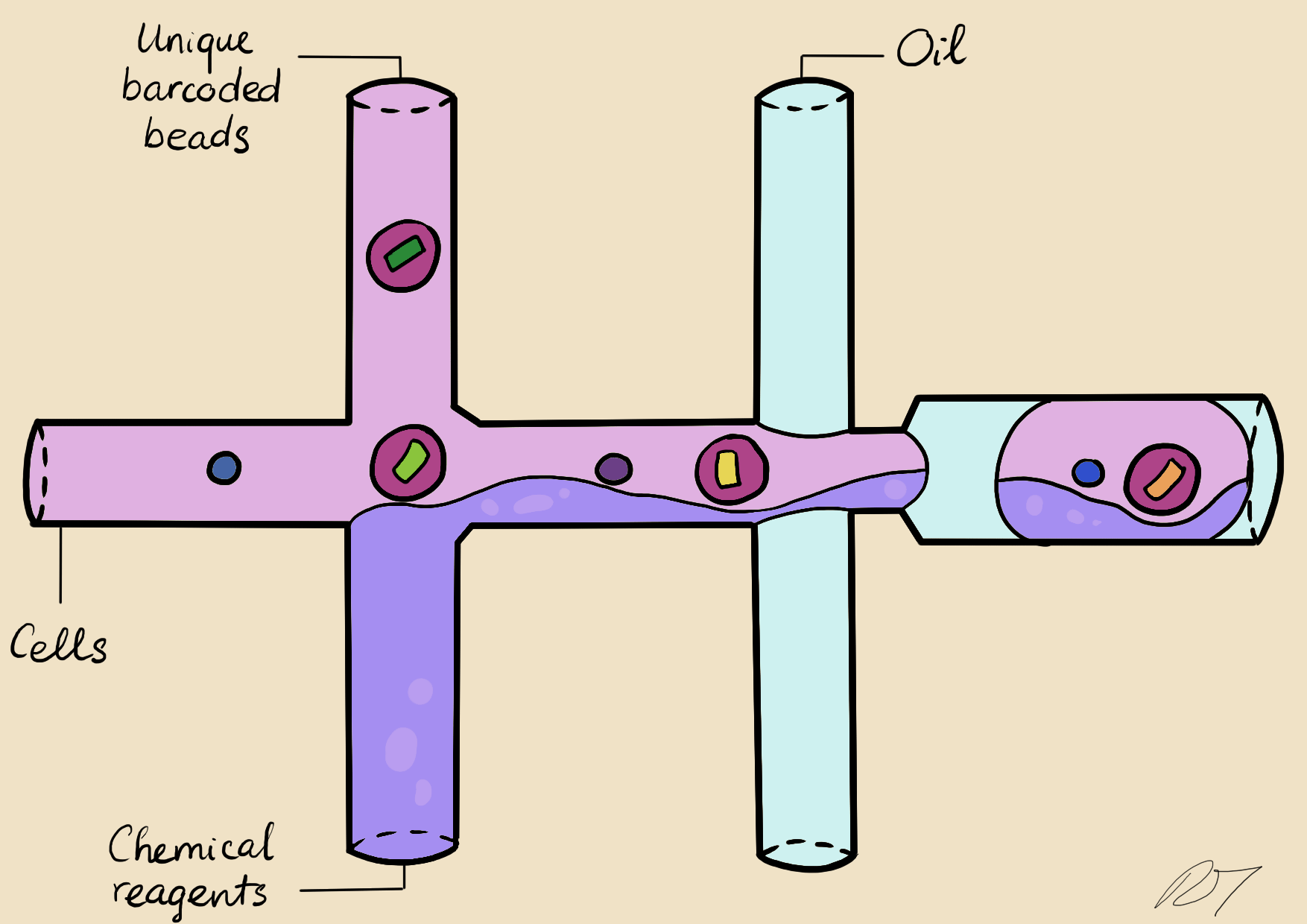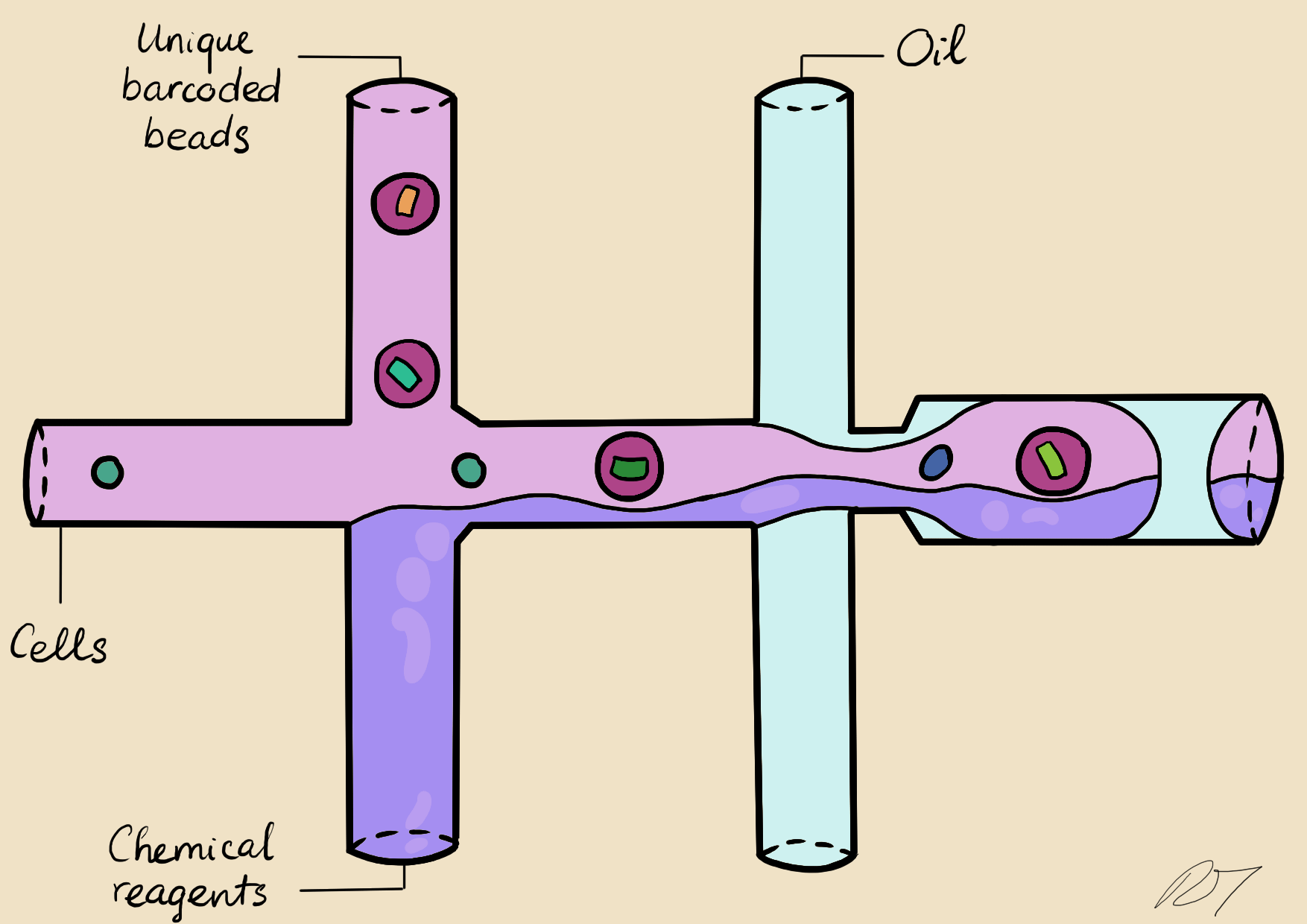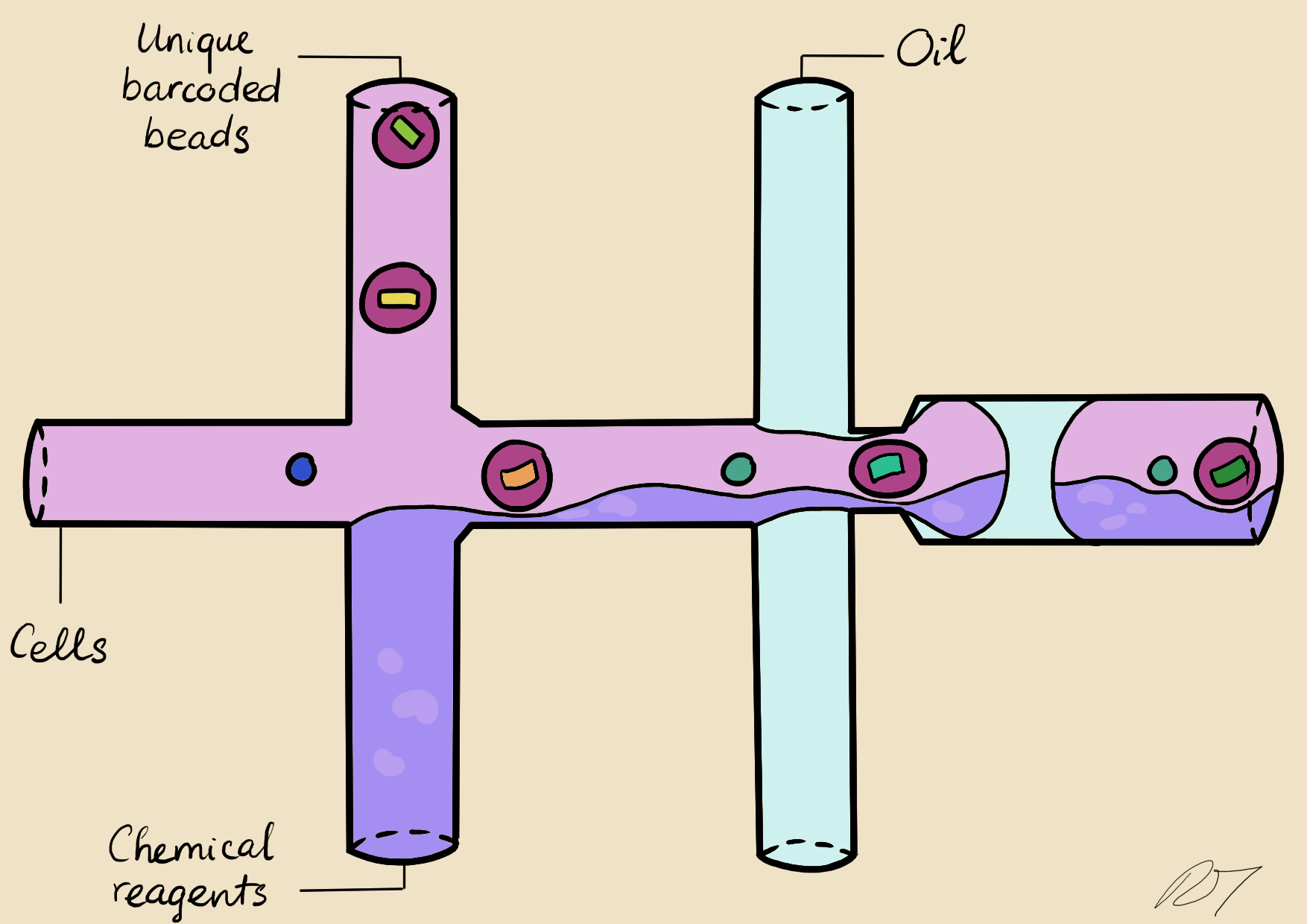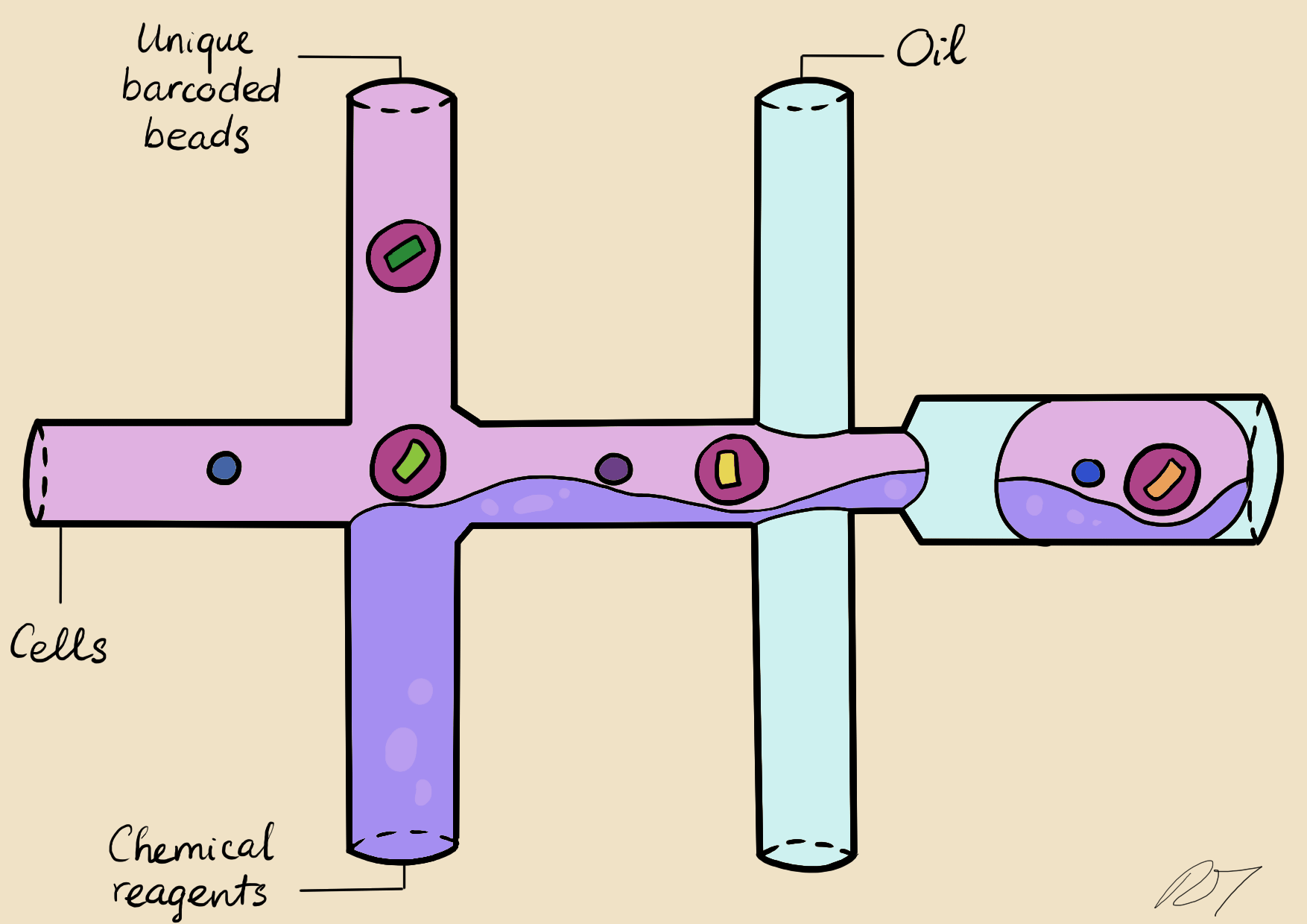
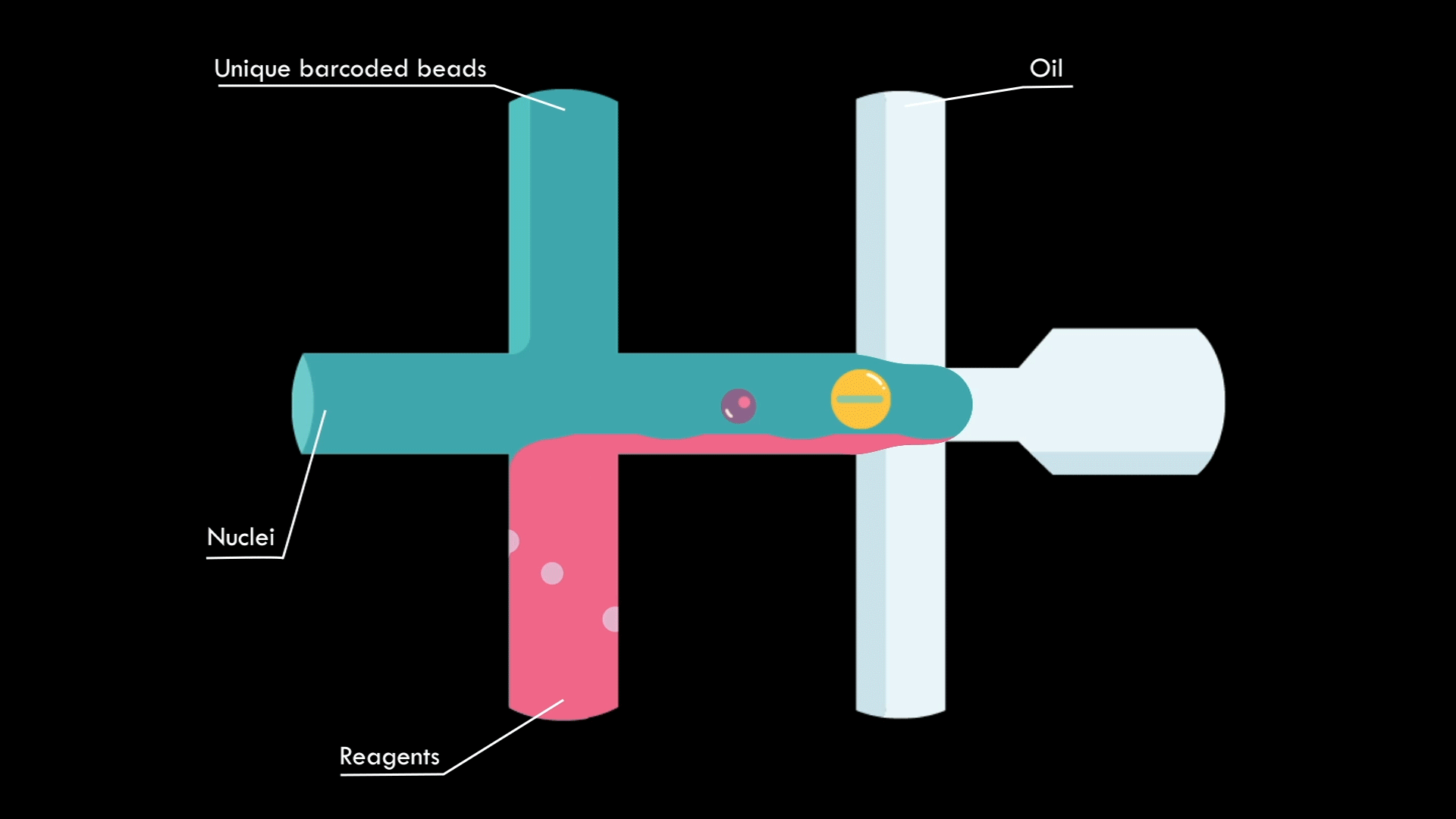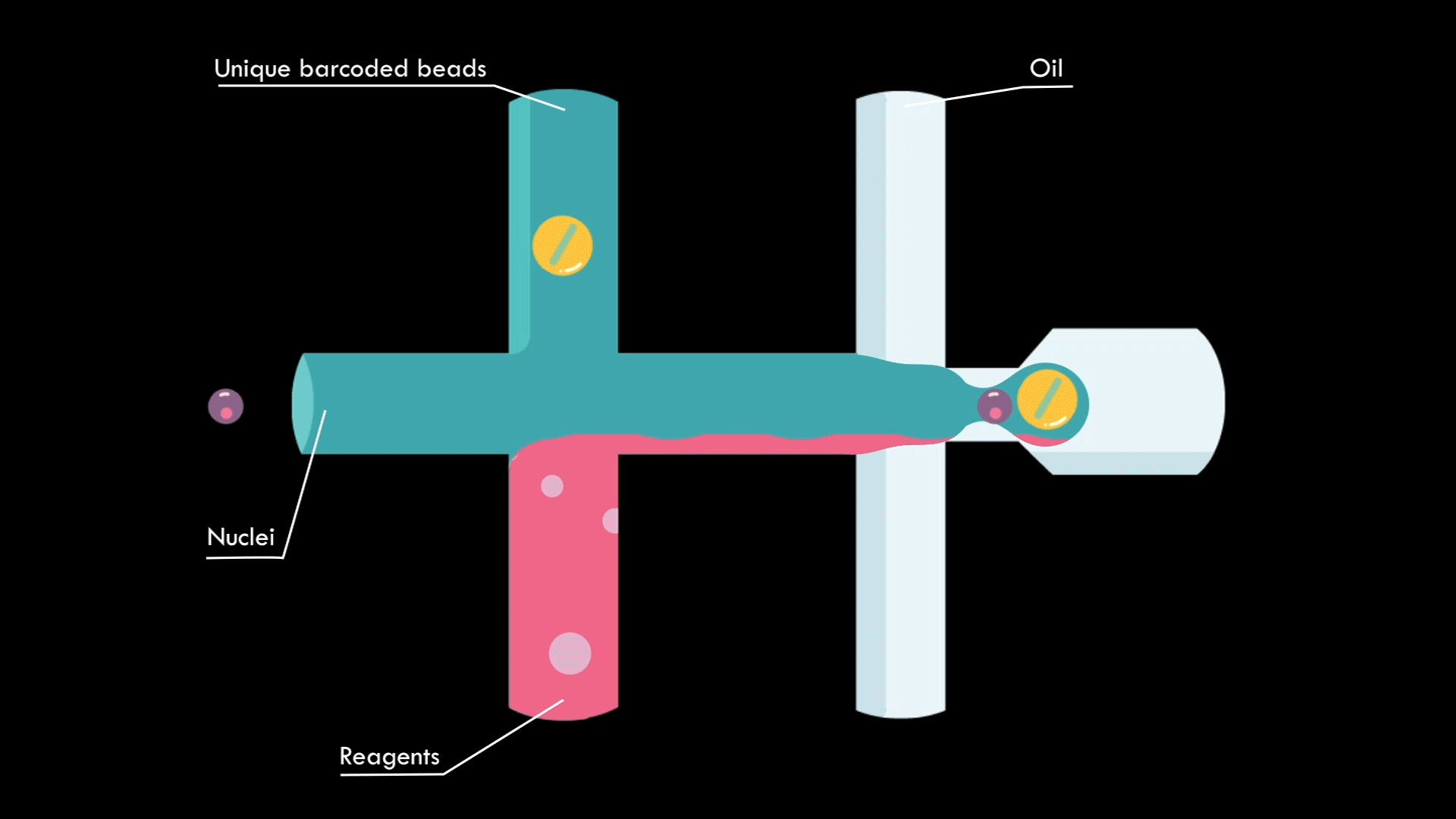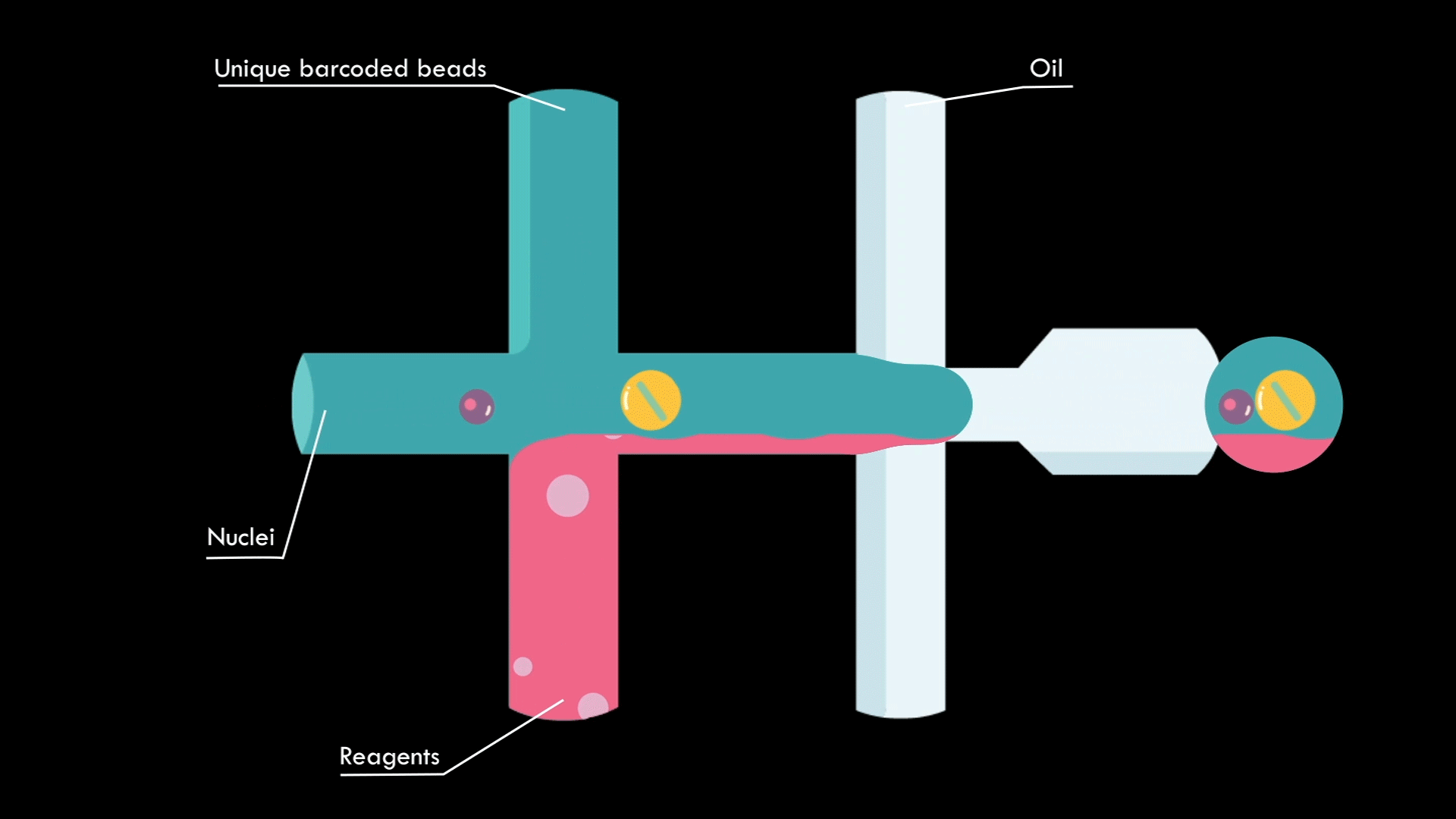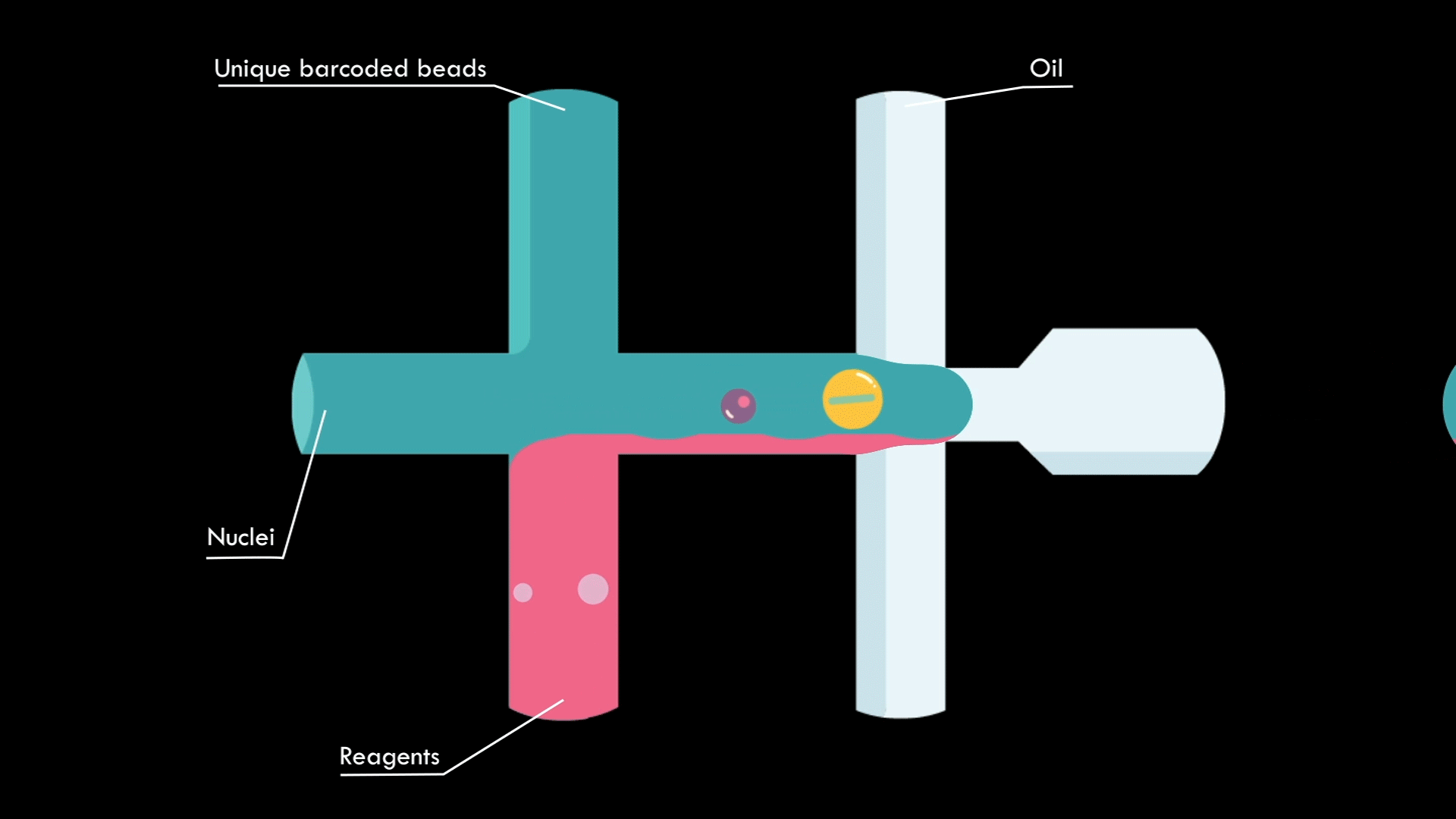
In a microfluidic system, individual nuclei are combined in a droplet along with an unique barcoded bead
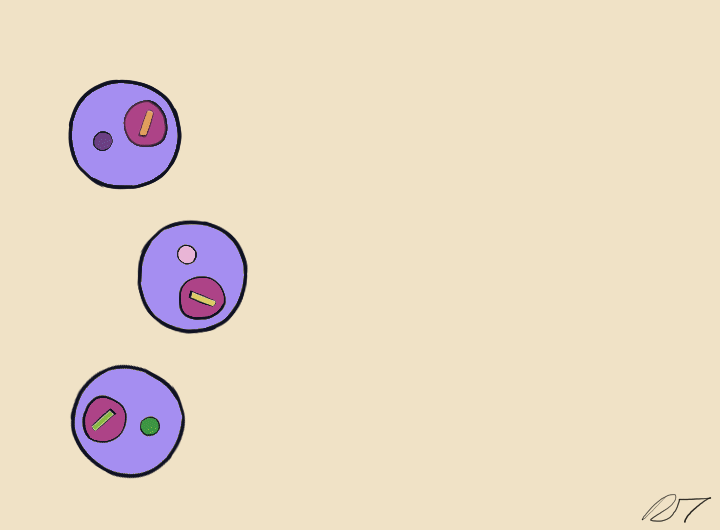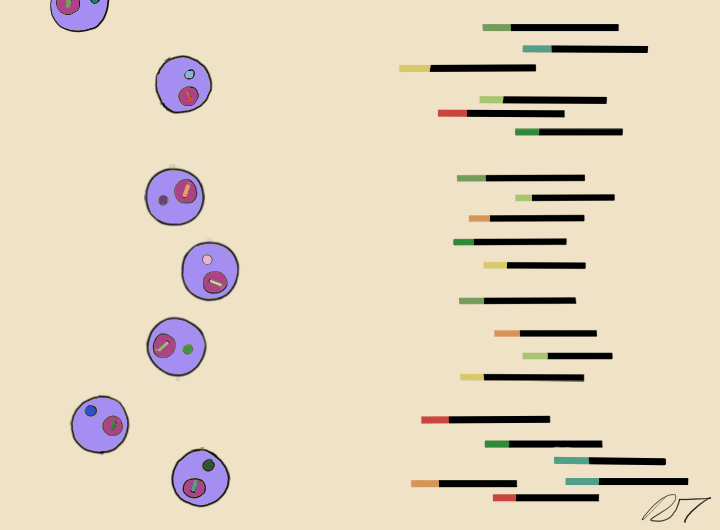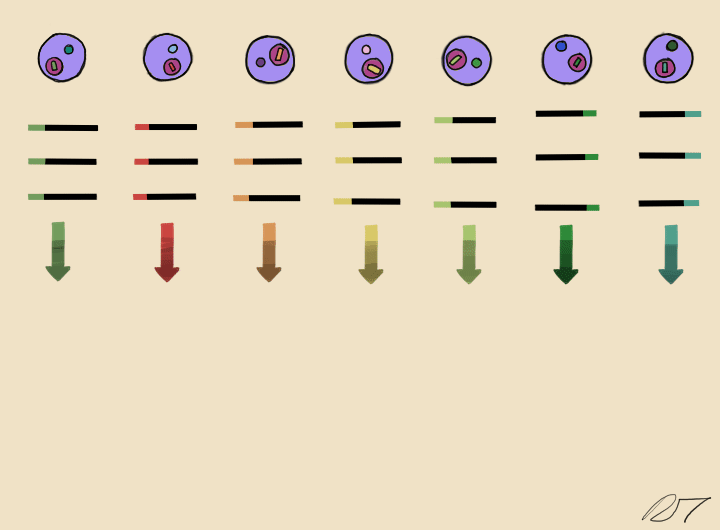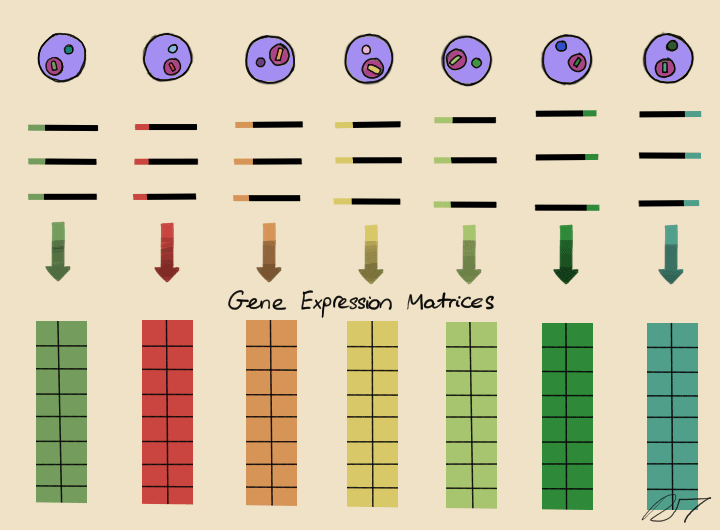
Droplets are pooled before cDNA libraries are generated and sequenced. Barcoded reads can then be traced back to the droplet and cell from which they originated. The transcriptomic profile of each cell is embedded in gene expression matrices.
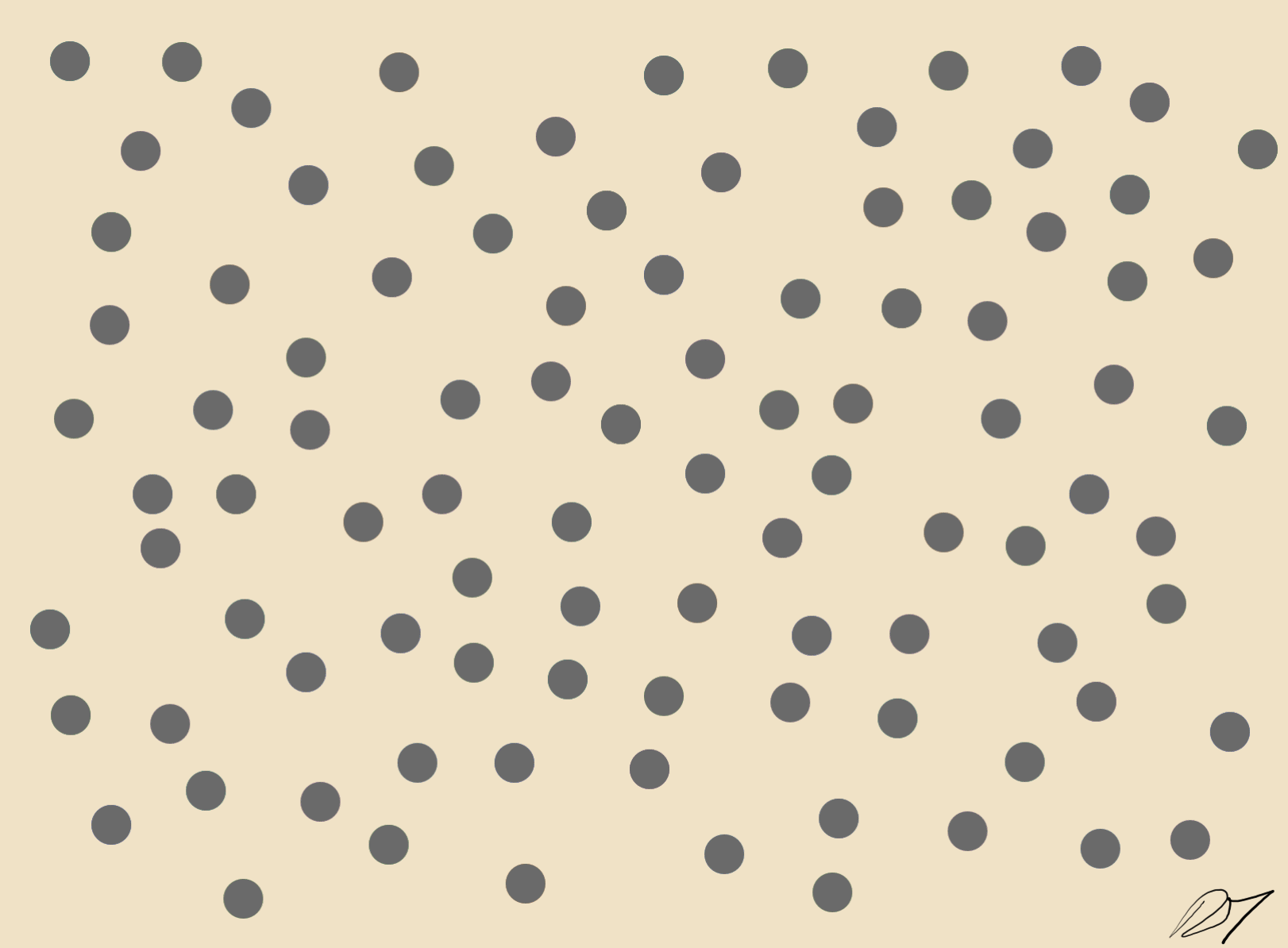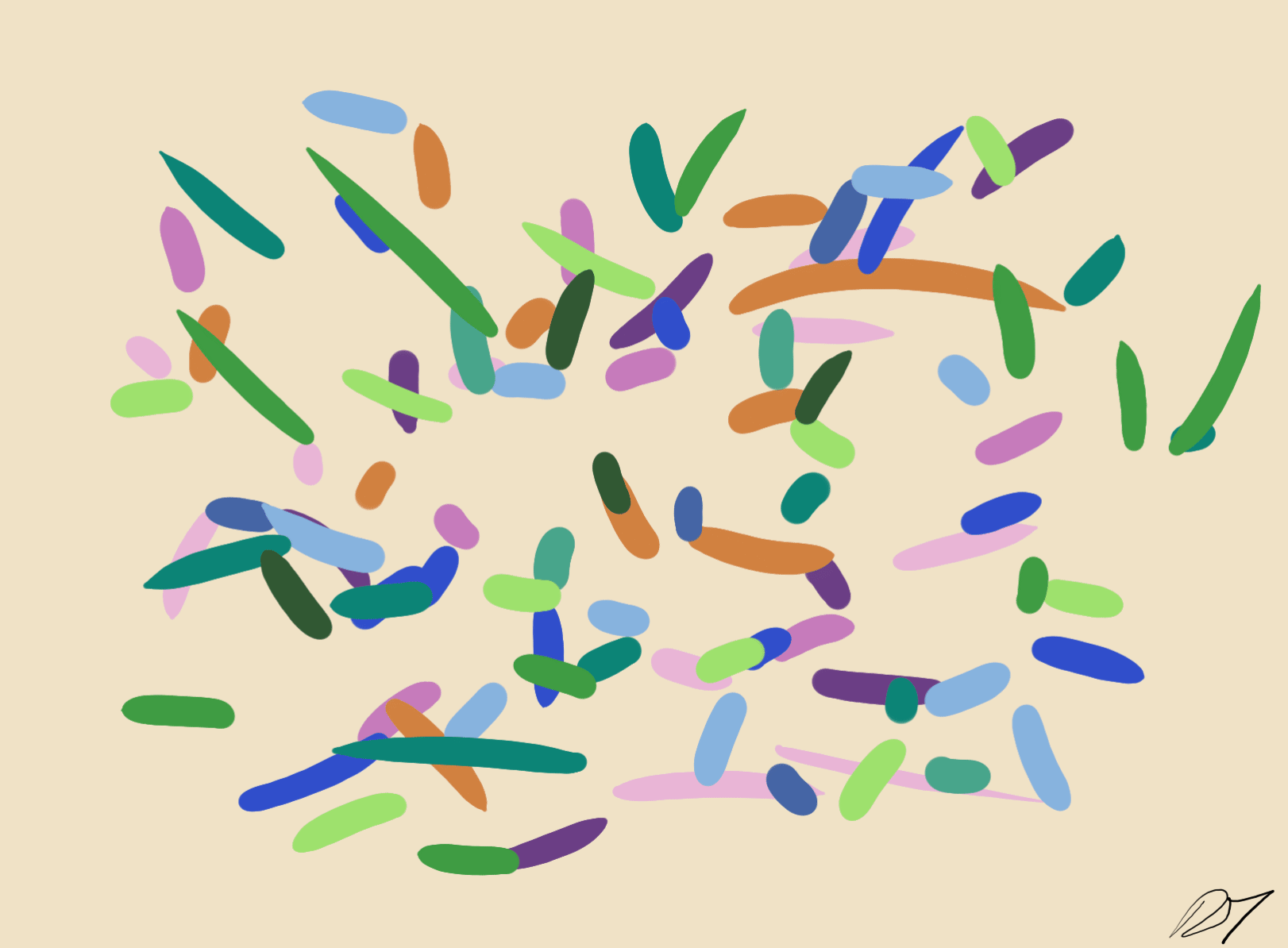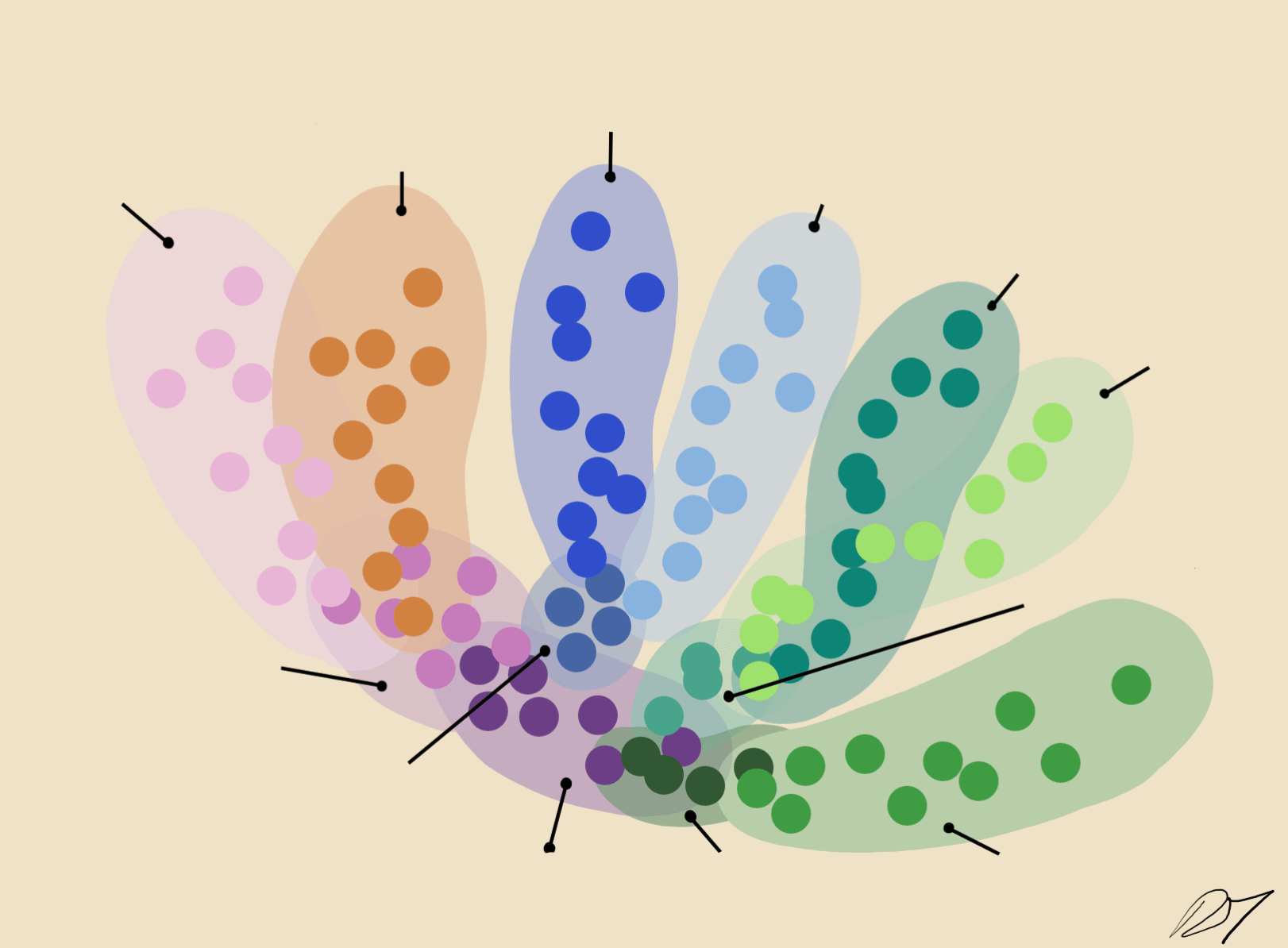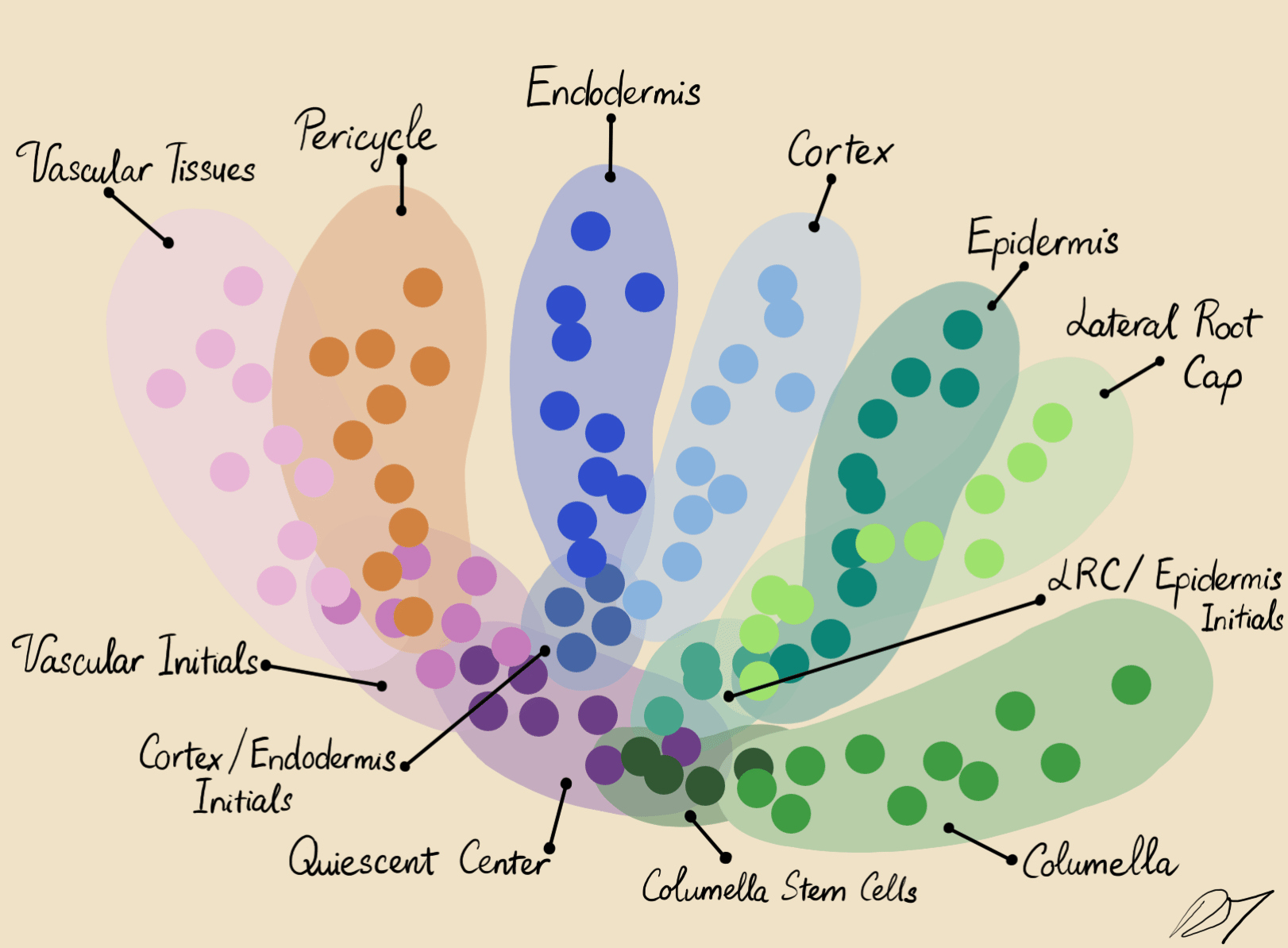
By comparing the similarities of gene expression matrices, corresponding cells can be sorted into clusters of distinguishable identities.

The same microfluidics system as above, but I'm learning to create animations in After Effects with assets from Illustrator!